This is the release note of v2.10.0.
Highlights
New CLI Subcommand for Analyzing Studies
New subcommands optuna trials, optuna best-trial and optuna best-trials have been introduced to Optuna’s CLI for listing trials in studies with RDB storages. It allows direct interaction with trial data from the command line in various formats including human readable tables, JSON or YAML. See the following examples:
Show all trials in a study.
$ optuna trials --storage sqlite:///example.db --study-name example
+--------+---------------------+---------------------+---------------------+----------------+---------------------+----------+
| number | value | datetime_start | datetime_complete | duration | params | state |
+--------+---------------------+---------------------+---------------------+----------------+---------------------+----------+
| 0 | 0.6098421143538713 | 2021-10-01 14:36:46 | 2021-10-01 14:36:46 | 0:00:00.026059 | {'x': 'A', 'y': 6} | COMPLETE |
| 1 | 0.6584108953598753 | 2021-10-01 14:36:46 | 2021-10-01 14:36:46 | 0:00:00.023447 | {'x': 'A', 'y': 10} | COMPLETE |
| 2 | 0.612883262548314 | 2021-10-01 14:36:46 | 2021-10-01 14:36:46 | 0:00:00.021577 | {'x': 'C', 'y': 3} | COMPLETE |
| 3 | 0.09326753798819143 | 2021-10-01 14:36:46 | 2021-10-01 14:36:46 | 0:00:00.024183 | {'x': 'A', 'y': 0} | COMPLETE |
| 4 | 0.7316749689191168 | 2021-10-01 14:36:46 | 2021-10-01 14:36:46 | 0:00:00.021994 | {'x': 'C', 'y': 4} | COMPLETE |
+--------+---------------------+---------------------+---------------------+----------------+---------------------+----------+Show the best trial as YAML.
$ optuna best-trial --storage sqlite:///example.db --study-name example --format yaml
datetime_complete: '2021-10-01 14:36:46'
datetime_start: '2021-10-01 14:36:46'
duration: '0:00:00.024183'
number: 3
params:
x: A
y: 0
state: COMPLETE
value: 0.09326753798819143Show the best trials of multi-objective optimization and train a neural network with one of the best parameters.
$ STORAGE=sqlite:///example.db
$ STUDY_NAME=example-mo
$ optuna best-trials --storage $STORAGE --study-name $STUDY_NAME
+--------+-------------------------------------------+---------------------+---------------------+----------------+--------------------------------------------------+----------+
| number | values | datetime_start | datetime_complete | duration | params | state |
+--------+-------------------------------------------+---------------------+---------------------+----------------+--------------------------------------------------+----------+
| 0 | [0.23884292794146034, 0.6905832476748404] | 2021-10-01 15:02:32 | 2021-10-01 15:02:32 | 0:00:00.035815 | {'lr': 0.05318673615579818, 'optimizer': 'adam'} | COMPLETE |
| 2 | [0.3157886300888031, 0.05110976427394465] | 2021-10-01 15:02:32 | 2021-10-01 15:02:32 | 0:00:00.030019 | {'lr': 0.08044012012204389, 'optimizer': 'sgd'} | COMPLETE |
+--------+-------------------------------------------+---------------------+---------------------+----------------+--------------------------------------------------+----------+
$ optuna best-trials --storage $STORAGE --study-name $STUDY_NAME --format json > result.json
$ OPTIMIZER=`jq '.[0].params.optimizer' result.json`
$ LR=`jq '.[0].params.lr' result.json`
$ python train.py $OPTIMIZER $LRSee #2847 for more details.
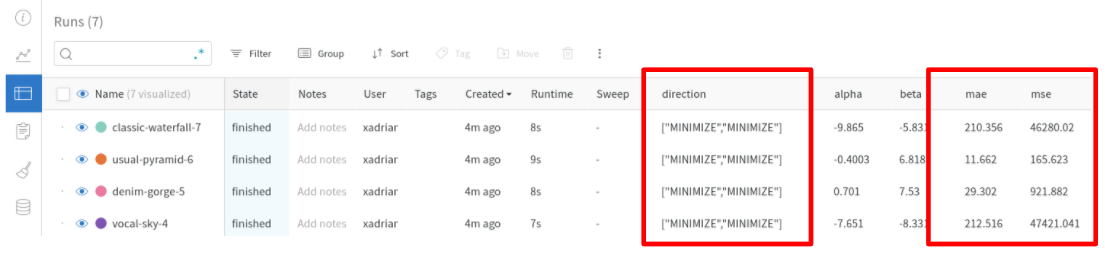
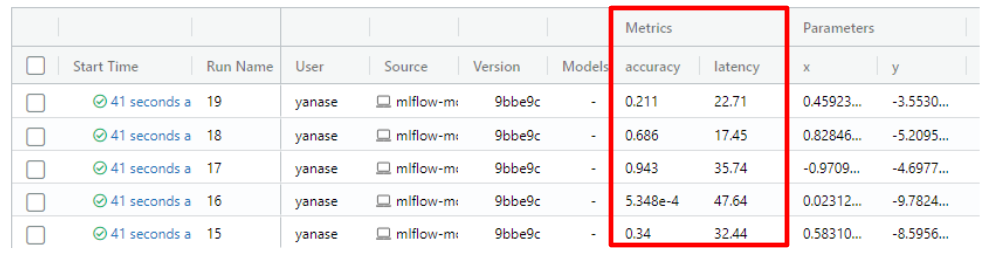
Multi-objective Optimization Support of Weights & Biases and MLflow Integrations
Weights & Biases and MLflow integration modules support tracking multi-objective optimization. Now, they accept arbitrary numbers of objective values with metric names.
Weights & Biases
from optuna.integration import WeightsAndBiasesCallback
wandbc = WeightsAndBiasesCallback(metric_name=["mse", "mae"])
...
study = optuna.create_study(directions=["minimize", "minimize"])
study.optimize(objective, n_trials=100, callbacks=[wandbc])MLflow
from optuna.integration import MLflowCallback
mlflc = MLflowCallback(metric_name=["accuracy", "latency"])
...
study = optuna.create_study(directions=["minimize", "minimize"])
study.optimize(objective, n_trials=100, callbacks=[mlflc])See #2835 and #2863 for more details.
Breaking Changes
- Align CLI output format (#2882)
- In particular, the return format of
optuna askhas been simplified. The first layer of nesting with the key “trial” is removed. Parsing can be simplified fromjq ‘.trial.params’tojq ‘.params’.
- In particular, the return format of
New Features
- Support multi-objective optimization in
WeightsAndBiasesCallback(#2835, thanks @xadrianzetx!) - Introduce trials CLI (#2847)
- Support multi-objective optimization in
MLflowCallback(#2863, thanks @xadrianzetx!)
Enhancements
- Add Plotly-like interpolation algorithm to
optuna.visualization.matplotlib.plot_contour(#2810, thanks @xadrianzetx!) - Sort values when the categorical values is numerical in
plot_parallel_coordinate(#2821, thanks @TakuyaInoue-github!) - Refactor
MLflowCallback(#2855, thanks @xadrianzetx!) - Minor refactoring of
plot_parallel_coordinate(#2856) - Update
sklearn.py(#2966, thanks @Garve!)
Bug Fixes
- Fix
datetime_completein_CachedStorage(#2846) - Hyperband no longer assumes it is the only pruner (#2879, thanks @cowwoc!)
- Fix method
untransformof_SearchSpaceTransformwithdistribution.single() == True(#2947, thanks @yoshinobc!)
Installation
- Avoid
keras2.6.0 (#2851) - Drop
tensorflowandkerasversion constraints (#2852) - Avoid latest
allennlp==2.7.0(#2894) - Introduce the version constraint of scikit-learn (#2953)
Documentation
- Fix
bounds' shape in the document (#2830) - Simplify documentation of
FrozenTrial(#2833) - Fix typo: replace CirclCI with CircleCI (#2840)
- Added alternative callback function #2844 (#2845, thanks @DeviousLab!)
- Update URL of cmaes repository (#2857)
- Improve the docstring of
MLflowCallback(#2883) - Fix
create_trialdocument (#2888) - Fix an argument in docstring of
_CachedStorage(#2917) - Use
:obj:forTrue,False, andNoneinstead of inline code (#2922) - Use inline code syntax for
constraints_func(#2930) - Add link to Weights & Biases example (#2962, thanks @xadrianzetx!)
Examples
- Do not use latest
keras==2.6.0(optuna/optuna-examples#44) - Fix typo in Dask-ML GitHub Action workflow (optuna/optuna-examples#45, thanks @jrbourbeau!)
- Support Python 3.9 for TensorFlow and MLFlow (optuna/optuna-examples#47)
- Replace deprecated argument
lrwithlearning_ratein tf.keras (optuna/optuna-examples#51) - Avoid latest
allennlp==2.7.0(optuna/optuna-examples#52) - Save checkpoint to tmpfile and rename it (optuna/optuna-examples#53)
- PyTorch checkpoint cosmetics (optuna/optuna-examples#54)
- Add Weights & Biases example (optuna/optuna-examples#55, thanks @xadrianzetx!)
- Use
MLflowCallbackin MLflow example (optuna/optuna-examples#58, thanks @xadrianzetx!)
Tests
- Fixed relational operator not including
1(#2865, thanks @Yu212!) - Add scenario tests for samplers (#2869)
- Add test cases for storage upgrade (#2890)
- Add test cases for
show_progress_barofoptimize(#2900, thanks @xadrianzetx!) - Speed-up sampler tests by using random sampling of skopt (#2910)
- Fixes
namedtupletype name (#2961, thanks @sobolevn!)
Code Fixes
- Changed y-axis and x-axis access according to matplotlib docs (#2834, thanks @01-vyom!)
- Fix a BoTorch deprecation warning (#2861)
- Relax metric name type hinting in
WeightsAndBiasesCallback(#2884, thanks @xadrianzetx!) - Fix recent
alembic1.7.0 type hint error (#2887) - Remove old unused
Trial._after_funcmethod (#2899) - Fixes
namedtupletype name (#2961, thanks @sobolevn!)
Continuous Integration
- Enable act to run for other workflows (#2656)
- Drop
tensorflowandkerasversion constraints (#2852) - Avoid segmentation fault of
test_lightgbm.pyon macOS (#2896)
Other
- Preinstall RDB binding Python libraries in Docker image (#2818)
- Bump to v2.10.0.dev (#2829)
- Bump to v2.10.0 (#2975)
Thanks to All the Contributors!
This release was made possible by the authors and the people who participated in the reviews and discussions.
@01-vyom, @Crissman, @DeviousLab, @Garve, @HideakiImamura, @TakuyaInoue-github, @Yu212, @c-bata, @cowwoc, @himkt, @hvy, @jrbourbeau, @keisuke-umezawa, @not522, @nzw0301, @sobolevn, @toshihikoyanase, @xadrianzetx, @yoshinobc